Protein stability prediction using deep learning
Protein stability prediction using deep learning
computational methods for separating oncogenic mutations from neutral mutations of the p53 protein using mathematical processing of the following calculated values for each sample
Machine learning model and deep learning was used to achieve correlations between calculated and experiment data
We select experimental data that are in the region of increased concentration values of the denaturant. Among the calculated data, we highlight two calculated parameters - this is the stability parameter lg(cond(W)), the so-called conditionality number, on the one hand, and the measure of entropy change, on the other hand. In this case, the measure of entropy change contains two areas - a positive area and a negative area
Correlation data
- We present a specific machine learning model to predict the stability of missense mutation in TP53 using the example of a combination of several physical experiments in which the unfolding of P53 mutations was studied depending on the denaturant concentration.
- The thermodynamic parameters of common missense mutations were determined by calculations and compared with those of the native p53 DNA-binding domain experiments.
- The effect of common cancer mutations on the thermodynamic stability of wild type p53 has been well studied by urea denaturation.
Comparison between experimental vs. calculated values
to which machine learning methods and clustering will be applied to develop a method for predicting the stability of mutant proteins
- lg(cond(W))The stability parameter is approximately equal to Kd.
- lg[Kd]calculated dissociation constant
-
m slope
experimental value - TdSthe differential entropy change
-
[Urea]50%, M
Denaturant concentration -
ddG(D-N)(H2O)
experimental value, or more precisely its change compared to the wild type
Thermodynamic stability of wild-type and mutant p53 core
How factors like temperature and pH affect a protein's stability
[Thermodynamic stability of wild-type and
mutant p53 core domain]
mutant p53 core domain]

[Semirational design of active tumor suppressor p53 DNA binding domain with enhanced stability]
[Mechanism of rescue of common p53 cancer mutations by second-site suppressor mutations]
[Structures of oncogenic, suppressor and rescued p53 core-domain variants: mechanisms of mutant p53 rescue]




p53 protein denaturation curves obtained in 4 different experiments
Changes in free energy of urea-induced unfolding of p53 core domain mutants
Equilibrium denaturation of p53 core domain
Comparison of experimental data from four studies

- Combined data from four experimental studies, extended data from which will be used in our study.
2.Let us dwell on experimental values that have the same sign for all studies.
3.This parameter is the denaturant concentration [Urea]50%, M
3.This parameter is the denaturant concentration [Urea]50%, M
Table of experimental values and calculated values for p53 mutations

T123A
V143A
H168R
G245S
R249S
V143A/N268D
G245S/N239Y
R249S/T123A
R249S/H168RT123A/H168R
R249S/T123A/H168R
V157FV157F/N235K
V157F/N235K/N239Y
R175HIS
C242SER
R248Q
R273H
Q104P
Q104HIS
A129D
A129E
A129S
M133L
D148E
D148S
T150P
Q165K
Q165E
R174K
C182S
L201P
V203A
L206S
D228E
N239Y
S260P
N268D
Q104HIS
A129D
A129E
A129S
M133L
D148E
D148S
T150P
Q165K
Q165E
R174K
C182S
L201P
V203A
L206S
D228E
N239Y
S260P
N268D
p53 mutations:
N239Y/N268D
M133L/V203A
M133L/N239T/N268D
V203A/N239Y/N268D
M133L/V203A/N239Y/N268D
Effects of Mutations on the Stability of T-p53C
The nature of the required dependencies are presented in the following diagrams.

Calculated values using our software
Experimental value
Experimental value
Corelation rate=0.9
Corelation rate=0.78
Corelation rate=-0.66
Corelation rate=-0.63
Corelation rate=0.62
Corelation rate=-0.67
The maximum correlation dependence for lg(cond(W))/[Urea]50%
The maximum correlation dependence between the calculated and experimental values under conditions of p53 protein denaturation was found between lg(cond(W)) and the denaturant concentration [Urea]50% in the region of increased concentrations required for denaturation of 50% of the protein in solution. Starting with C1=2.8M and more.
The correlation dependence between the values reached 90%
The correlation dependence between the values reached 90%
The maximum correlation dependence for TdS/[Urea]50%
The maximum correlation dependence between the calculated and experimental values under conditions of p53 protein denaturation was found between TdS and the denaturant concentration [Urea]50% in the region of increased concentrations required for denaturation of 50% of the protein in solution. Starting with C1=2.8M and more.
List of p53 protein mutations, the denaturation of which requires an increased concentration of [Urea]50% denaturant
Correlation between calculated lg(cond(W)) and experimental data [urea]50% at the required increased denaturant concentration


p53 mutations
Q104P
Q104HIS
A129S
M133L
T150P
R174K
C182S
L201P
V203A
L206S
N239Y
S260P
N268D
Q104P
Q104HIS
A129S
M133L
T150P
R174K
C182S
L201P
V203A
L206S
N239Y
S260P
N268D
the concentration of denaturant exceeds 2.8 M
Dependencies between calculated and experimental data taking into account denaturant concentration.


I
II
Various physical quantities should be taken for subsequent analysis of the correlation between experimental and calculated data.

TdS>0
TdS<-1
TdS>0
TdS<-1
Entropy change
Stability change

Features of the correlation between the ddG experimental value and the calculated Kd, note that we take the logarithm of the dissociation constant, so the value can go into the negative region
Calculated value
Calculated value
Experimental value
Application of machine learning methods for data clustering

Selection of segments for correlation analysis

M133L 3.275 0.0315
D148E 3.04 0.0602
D148S 3.28 0.0963
Q165K 2.695 0.0407
V203A 3.345 0.0440
D228E 3.22 0.0603
N239Y 3.265 0.02665
M133L/V203A 3.405 0.075
M133L/V203A/N239Y/N268D
T123A 3.125 0.036149
H168R 2.285 0.104794
R249S/H168R2.6150.0542
R249S/T123A/H168R
V157F/N235K 2.54 0.097
V157F/N235K/N239Y
N239Y(aver)3.55 0.026
D148E 3.04 0.0602
D148S 3.28 0.0963
Q165K 2.695 0.0407
V203A 3.345 0.0440
D228E 3.22 0.0603
N239Y 3.265 0.02665
M133L/V203A 3.405 0.075
M133L/V203A/N239Y/N268D
T123A 3.125 0.036149
H168R 2.285 0.104794
R249S/H168R2.6150.0542
R249S/T123A/H168R
V157F/N235K 2.54 0.097
V157F/N235K/N239Y
N239Y(aver)3.55 0.026
Creativity is to discover a question that has never been asked. If one brings up an idiosyncratic question, the answer he gives will necessarily be unique as well.
Q104H 3.25 5.804
A129D 2.74 5.807
A129E 2.66 5.800
A129S 2.93 5.800
Q165E 2.53 5.801
C182S 3.06 5.797
N268D 3.505 5.806
R249S 2.625 5.802
R175HIS 2.265 5.800
C242SER 2.295 5.791
R273H 3.175 5.800
R249S(aver) 2.625 5.802
N268D(aver)3.53 5.806
A129D 2.74 5.807
A129E 2.66 5.800
A129S 2.93 5.800
Q165E 2.53 5.801
C182S 3.06 5.797
N268D 3.505 5.806
R249S 2.625 5.802
R175HIS 2.265 5.800
C242SER 2.295 5.791
R273H 3.175 5.800
R249S(aver) 2.625 5.802
N268D(aver)3.53 5.806
Various clustering methods used to automatically divide the resulting sample of points.
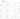
as can be seen from the graphs, various physical calculation data should be used to predict experimental data in the positive region of entropy change TdS;
in the region near zero we do not present correlation graphs;
in the negative region of entropy change, the calculated value characterizing stability lg(cond(W)) rather than disorder should be used
in the region near zero we do not present correlation graphs;
in the negative region of entropy change, the calculated value characterizing stability lg(cond(W)) rather than disorder should be used
positive region of the entropy change
area near zero
negative region of entropy change
Analysis of various areas of stability lg(cond(W)) of calculated data
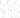
lg(cond(W))>lg(cond(W))wt
lg(cond(W))<lg(cond(W))wt
1
2

