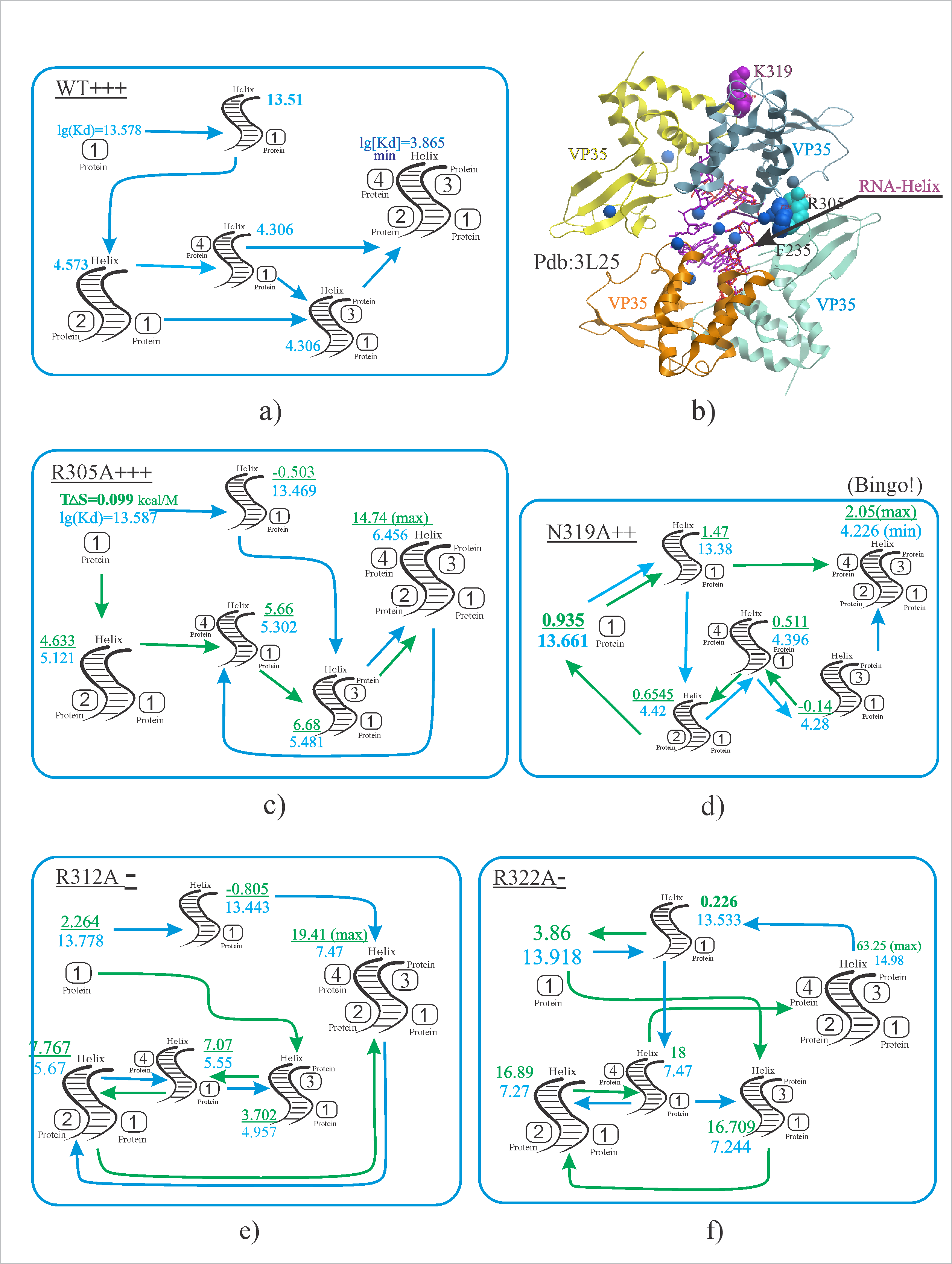
In what order will the hexamer assemble?
Identify viral protein interactions order during replication and transcription processes
Determining the sequence or molecules queues entering chemical reactions involves understanding the order in which reactants interact to form products. This process is crucial in reaction kinetics, enzymatic pathways, and drug
design. Below are the main approaches to determine these molecular queues.
In this study, we will analyze the order of complex molecular formations involving viral and human proteins, as well as RNA molecules.
The objects of study will include single-stranded and double-stranded RNA molecules, as well as small domains of viral proteins.

What are the proteins binding to RNA?
be calculated.
-
Calculations
Calculation of thermodynamic parameters for each bioformation -
Analysis
Analysis of the obtained data in the order of formation of the complex

lg(cond(W)), ∆(∆W), T∆S,
calculated using computational methods.According to experimental studies, the 8-bp dsRNA·LGP2 complex exhibited an estimated molecular mass consistent
with a 2:1 complex, while the hairpin RNA·LGP2 complex had a molecular mass consistent with a 1:1 complex. Assuming the simultaneous existence of several biological systems in solution, including LGP2 and dsRNA independently, as well as
[LGP2-dsRNA] and [LGP2-dsRNA-LGP2], the relative proportions of these systems may vary depending on experimental conditions such as reagent and salt concentrations, temperature, and the presence of modified proteins.
In this study, we calculate thermodynamic parameters for each compound and analyze energy barriers for transitions between the following biochemical systems:
• [LGP2]: monomer
• [8dsRNA]: double-stranded RNA
• [LGP2-8dsRNA]: 1:1 complex
• [LGP2-8dsRNA-LGP2]: 2:1 complex
Lower values of lg(cond(W)) indicate a more stable biological compound closer to thermodynamic equilibrium. Conversely, higher lg(cond(W)) values correspond to less stable compounds further from equilibrium, implying a system in search of further chemical reactions to reach equilibrium.
Theoretical approach for determining the number of subunits (monomers) in a multimolecular complex
For each molecular object, a spectrum of calculated thermodynamic parameters will be determined, such as the stability parameter, the measure of differential entropy change, and the change in energy .As additional monomers attach to a monomer and the structure becomes more complex, the set of calculated physical parameters will also change. Therefore, we will create a table outlining all possible pathways for forming the final complex molecular assembly and calculate their thermodynamic parameters (see Table 1).

[LGP2] monomer (a), [LGP2-8dsRNA] complex 1:1 (b), and [LGP2-8dsRNA-LGP2] complex 2:1 (c). Combined calculated graphs for the three molecular formations are presented for the values of ∆(∆W) (d), T∆S (e), and lg(cond(W)) (f).
In Fig.4 shows the calculated results of the stability value lg(cond(W)) for different
types of intermediate reactions:
a significant reduction in stability, as observed in the case of mut[LGP2] E573A. The remaining three monomeric complexes—mut[LGP2] K634E, mut[LGP2] K651E, and mut[LGP2] K634E/R636E—show a sharp decline in the values of log(cond(W)). This trend of increased instability at the very beginning of the biological complex formation at the monomeric protein stage may indicate that the thermodynamic equilibrium of the monomeric biological system is reached even before interaction with 8dsRNA.



various substitutions (E573, I597, K634, R636, K634, K651) in the amino acid sequence.
• Figure 4b presents the calculated value of lg(cond(W)) for the [LGP2-RNA] biological
formation. 168
• Figure 4c shows the calculated lg(cond(W)) value for the [LGP2-RNA-LGP2] complex.
• Figure 4d provides information on the changes in dissociation energy for the three
biological formations: [LGP2], [LGP2-RNA], and [LGP2-RNA-LGP2], indicating the
direction and numbering of intermediate reactions. 172
• Figure 4e depicts changes in entropy for the studied biological formations: [LGP2],
[LGP2-RNA], and [LGP2-RNA-LGP2].
The comparison between Fig. 4d) and Fig. 4e reveals unsynchronized changes in
the calculated physical properties of dissociation energy and differential entropy. We will
summarize the conclusions and results in a final Table 4-6


mediate complexes, such as mutant LGP2 monomers and the [LGP2-8dsRNA] complex
(1:1). The mutant monomer (mutLGP2) was shown to be more stable compared to the
wild-type protein. Upon binding to dsRNA, the [LGP2-8dsRNA] complex exhibited even
higher stability, reflected by lower values of lg(cond(W)).
be calculated.
-
Calculations
Calculation of thermodynamic parameters for each bioformation -
Analysis
Analysis of the obtained data in the order of formation of the complex
be calculated.
-
Calculations
Calculation of thermodynamic parameters for each bioformation -
Analysis
Analysis of the obtained data in the order of formation of the complex

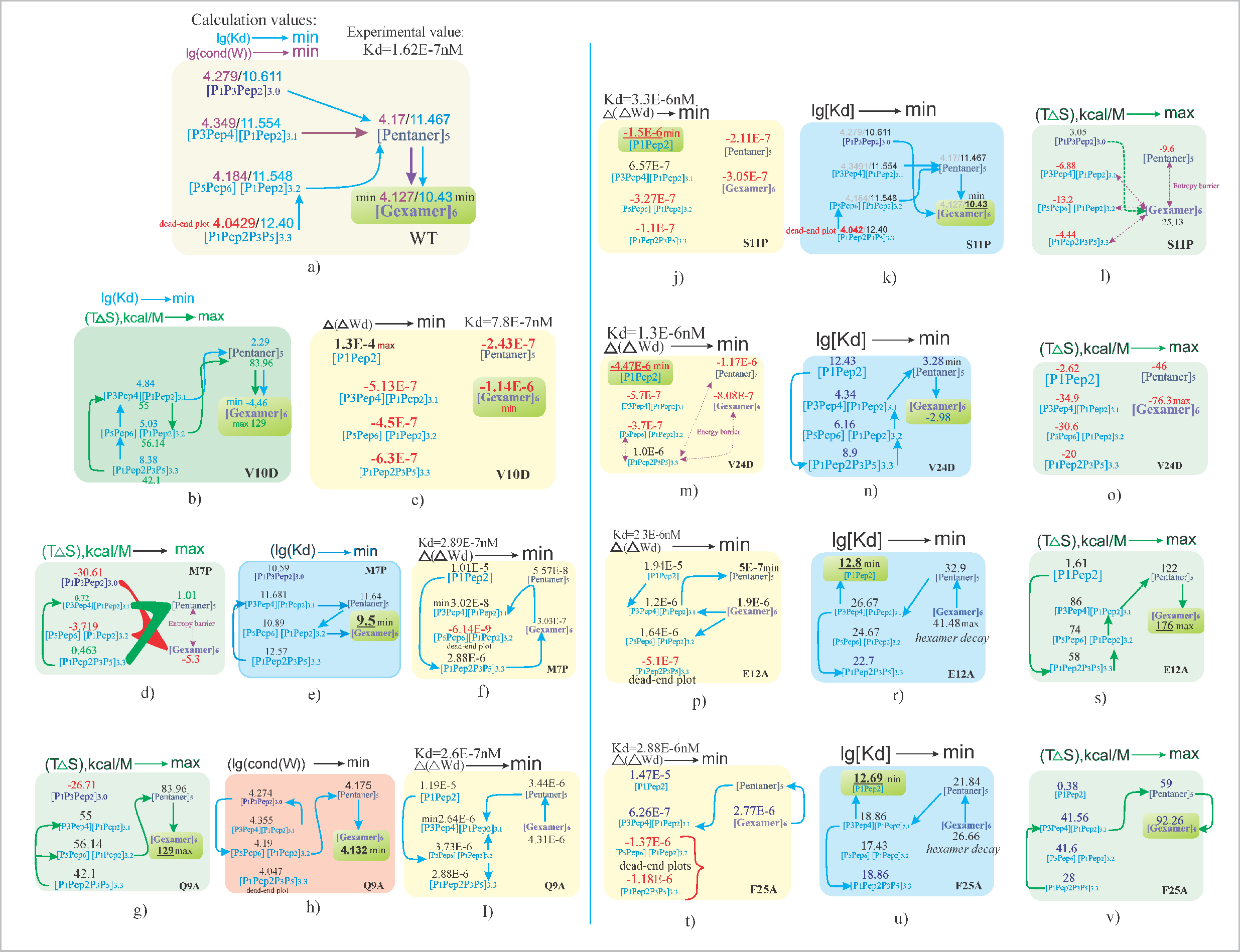
Scheme of chemical reaction pathways on the assembly of a hexamer, taking into account the change in the calculated thermodynamic quantities T∆S, lg(Kd), ∆(∆W), and lg(cond(W)).
The detailed analysis of hexamer assembly pathways reveals critical insights into the role of thermodynamic parameters T∆S, lg(Kd ), ∆(∆W), and lg(cond(W)) in guiding the formation of stable biochemical complexes. The study demonstrates that pathways
generally progress from states with high stability and affinity values toward a final hexamer
characterized by lower lg(cond(W)) and lg(Kd) values.