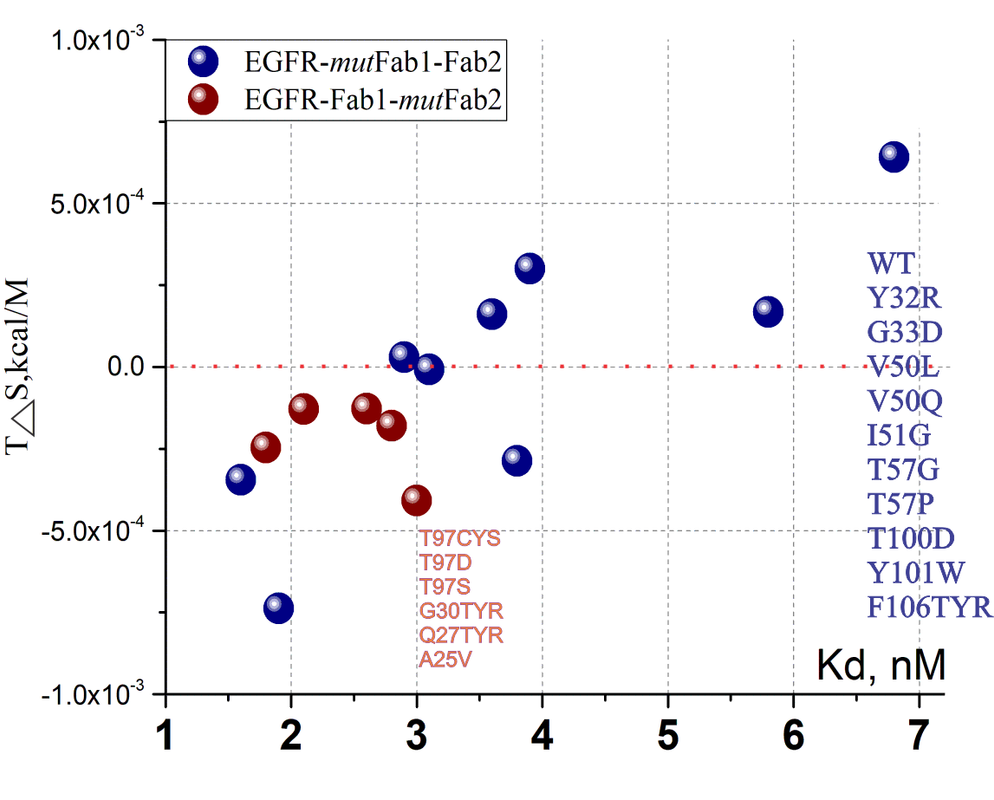
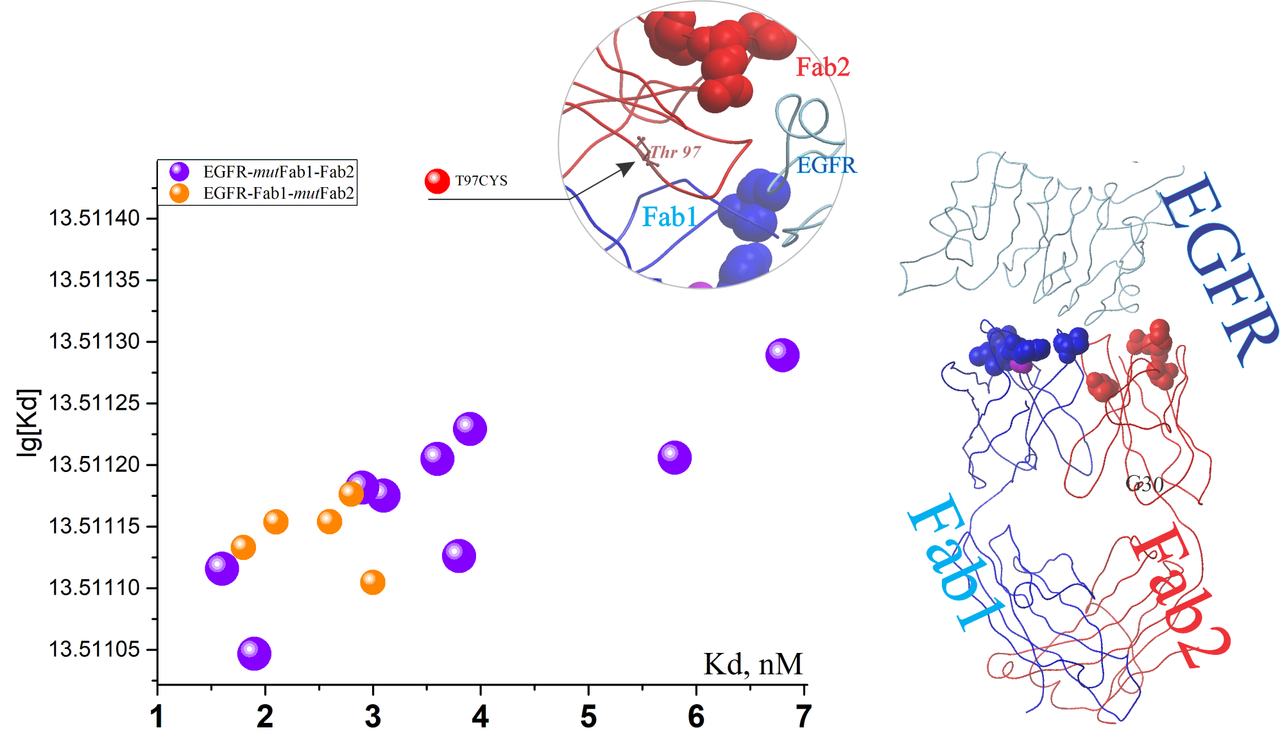
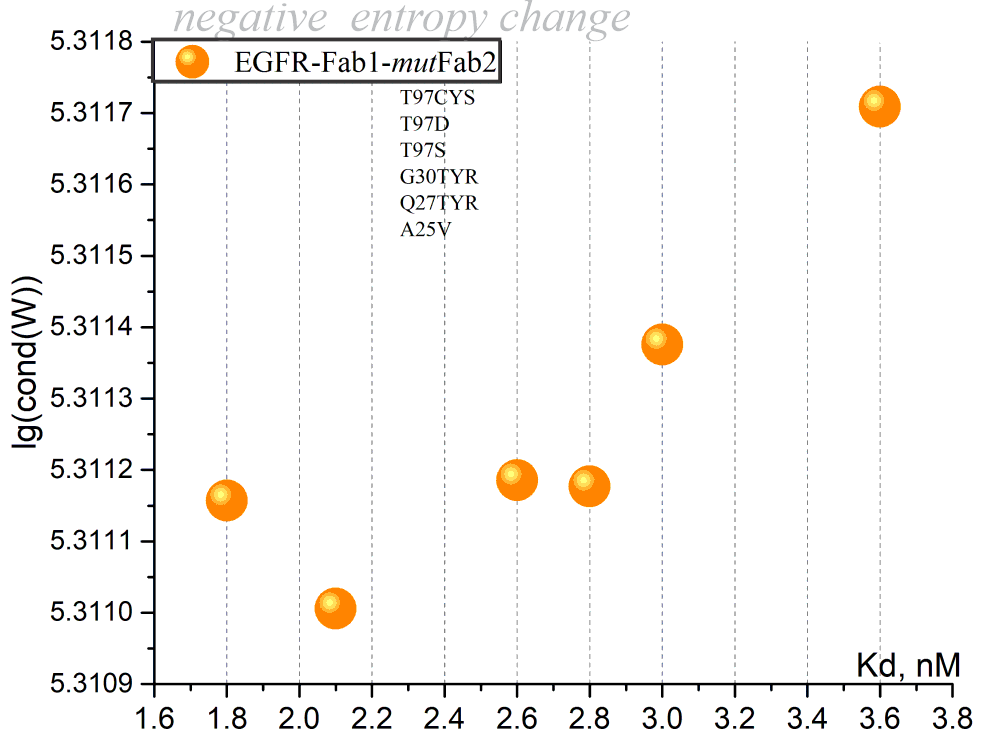
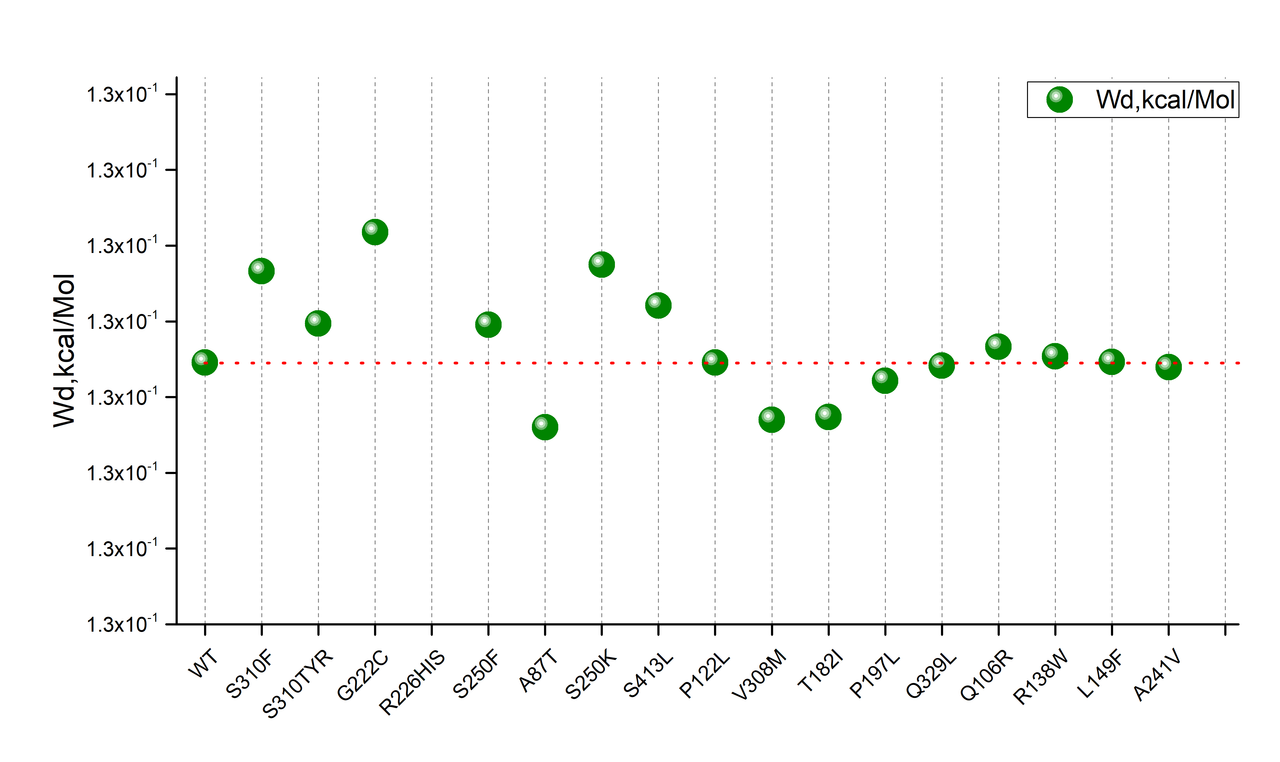
Deep mutational scanning of an antibody against EGFR
| Antibody Name | Target Antigen | UniProt ID | PDB Structures | IC₅₀ (nM) | Mutation Profile | Bioavailability | Preclinical Cost (USD) | Production Cost per 100mg (98%) | Study Reference |
|---|---|---|---|---|---|---|---|---|---|
| antibody 225 to create hu225. #1 | EGFR | P04626 | 1YY9 | nM | T97CYS,T97D, T97S, G30TYR, Q27TYR, A25VL | NA | NA | NA0 | Study Link |
FDA-Approved Therapeutic Monoclonal Antibodies with Expandable Details
| Antibody Name | Target Antigen | UniProt ID | PDB Structures | IC50 (nM) | Mutation Profile | Bioavailability | Preclinical Cost (USD) | Production Cost per 100mg | Study Reference | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Trastuzumab #1 (Click to expand details)
| |||||||||||||||||||||||||||
Trastuzumab drug response.
A panel of 12 parental and derived BCCLs were tested for trastuzumab response.
| Cell Line | Cell Viability (%) | SD | Growth Rate (ΔGR) | Response |
|---|---|---|---|---|
| HCC1419 | 83.97 | 0.08 | 1.15 | R |
| HCC1596 | 117.52 | 0.09 | 0.82 | R |
| HCC1954 | 101.14 | 0.02 | 0.94 | R |
| JIMT-1 | 82.60 | 0.08 | 1.11 | R |
| BT-474 | 38.70 | 0.07 | 3.52 | S |
| BT-474.R | 85.31 | 0.11 | 1.14 | R |
| SK-BR-3 | 50.67 | 0.05 | 2.13 | S |
| SK-BR-3.R | 86.25 | 0.02 | 1.11 | R |
| AU-565 | 72.15 | 0.04 | 1.26 | S |
| AU-565.R | 93.96 | 0.06 | 0.91 | R |
| EFM-192A | 60.00 | 0.17 | 1.73 | S |
| EFM-192A.R | 98.00 | 0.10 | 0.85 | R |
Low viability (eg <70%)
High growth reduction (ΔGR > 1.2)
E.g. BT-474, SK-BR-3
R (Resistant) — cells are resistant:
High viability (>80%), growth is preserved (ΔGR ≈ 1 or <1)
Often these are either initially resistant lines or “.R” — resistant derivatives.
Lapatinib in the management of breast cancer
The 2023 “Human Epidermal Growth Factor Receptor 2 (HER2) Breast Testing Guideline Update” reaffirms the 2018 “HER2 Breast Testing Guideline Focused Update.”
This reaffirmation was propelled by results of the 2022 DESTINY-Breast04 trial, which prompted the United States Food and Drug Administration (FDA) to expand the approval of the HER2 antibody-drug conjugate, trastuzumab deruxtecan, from metastatic breast cancer patients with HER2 protein over-expression/amplification to also include metastatic patients with HER2 IHC 1+ or 2+/ISH negative results. This guideline update does not support the use of a HER2-Low interpretive category because the DESTINY-Breast04 trial did not include patients with HER2 IHC 0 results, there is no evidence to support that IHC 1+ or 2+/ISH negative results are predictive of trastuzumab deruxtecan treatment response when compared to IHC 0 results. Therefore, the FDA expansion of approval to this group was only based on clinical trial eligibly criteria rather than a new predictive indication for HER2 testing.
The guideline update is published as an early online release in the Archives of Pathology & Laboratory Medicine.
A high level of HER2 gene amplification was proposed as improved predictor of sensitivity to trastuzumab treatment, but the studies defined different thresholds Furthermore, a tumour mutational burden (TMB) of more than 10 mutations per Mb was indicated to be a potential biomarker for trastuzumab efficacy ]. Moreover, initial resistance to trastuzumab or impaired treatment response was associated with the presence of co-amplifications and mutations of oncogenes or genes related to the HER2 signalling pathway such as EGFR and KRAS In addition, intratumoural heterogeneity of HER2 protein expression, frequently found in GC and GEJC, was associated with poor prognosis
In 2021, immunotherapy offered new treatment options for mGC/mGEJC patients and the combination of nivolumab/pembrolizumab and chemotherapy was approved as first-line therapy for programmed cell death ligand 1 (PD-L1) expressing tumours. Since then, PD-L1 expression is routinely assessed in clinical diagnostics in addition to the HER2 status []. Recent studies like the KEYNOTE-811 explored the efficacy of anti-PD-L1 treatment in HER2-positive mGC/mGEJC showing that the combination of pembrolizumab and trastuzumab with chemotherapy leads to improved response rates compared to treatment with chemotherapy and trastuzumab alone ]. In addition, the combined expression of PD-L1 and HER2 was found to be a positive prognostic factor for survival in GC Findings from a preliminary biomarker analysis of the DESTINY-Gastric03 trial showed a large overlap between HER2 and PD-L1 expression in 85% of the patients GC or GEJC In the KEYNOTE-811 study, similar frequencies for double positivity (HER2+/PD-L1+) have been reported
In this retrospective study, we divided HER2-positive mGC/mGEJC patients into two groups based on progression-free survival (PFS) under trastuzumab-based treatment and performed an explorative analysis of biomarkers. We applied targeted next-generation sequencing (NGS) using a 409 gene panel, Affymetrix gene expression analysis and immunohistochemistry to evaluate biomarkers and investigated their prognostic impact in this cohort.. In one study, response rates and time to progression were maintained from first to beyond second line treatment. Response rates were 42.6% in first line trastuzumab therapy, 25.9% in second line, and 30% in beyond second line respectively. Median time to progression was 6 months in all treatment lines
| Trial | Population | Treatment arms | Conclusion |
|---|---|---|---|
| 2006–2013 EGF105485 (TEACH) | HER2-positive, trastuzumab-naive patients who have completed adjuvant therapy | Lapatinib vs Placebo | DFS, did not reach the pre-specified statistical significance level |
| A Randomized, Double-blind, Multicenter, Placebo-controlled Study in Women with Early-Stage ErbB2 Overexpressing Breast Cancer | |||
| Hypothesis: oral single-agent lapatinib was expected to improve disease-free survival (DFS), but statistical significance was not reached. | |||
| Trial | Population | Treatment Arm | Response | ||
|---|---|---|---|---|---|
| Overall Resp. | No Change | Prog. Disease | |||
| 2008–2016 EGF105485 (ALTTO) | HER2-positive, adjuvant setting. Women with primary ErbB2 overexpressing and/or gene amplified breast cancer > 2 cm diameter who have not undergone previous treatment for invasive breast cancer | Lapatinib + Trastuzumab (N=152) | 102 | 45 | 5 |
| Lapatinib + Paclitaxel (N=154) | 81 | 33 | 2 | ||
| Trastuzumab + Paclitaxel (N=149) | 81 | 57 | 11 | ||
| Trial | Population | Treatment Arm | Overall Response Rate | At Surgery |
|---|---|---|---|---|
| 2008–2010 EGF106903 (Neo-ALTTO) | HER2-positive, neo-adjuvant setting. After surgery, all subjects were to receive three courses of adjuvant 5-fluorouracil, epirubicin and cyclophosphamide | Trastuzumab + Paclitaxel | 45% | 70% |
| Lapatinib | 52% | 74% | ||
| Lapatinib + Trastuzumab + Paclitaxel | 67% | 80% | ||
| At the time of surgery in the ITT population, response was statistically significantly higher in subjects receiving lapatinib plus trastuzumab with paclitaxel. | ||||
| Trial | Population | Treatment Arms | Drugs | Conclusion |
|---|---|---|---|---|
| GeparQuinto (GBG) | HER2-positive breast cancer, neoadjuvant Stages I–III (including inflammatory) | EC-DT = Epirubicin + Cyclophosphamide (4 cycles) → Docetaxel + Trastuzumab EC-DL = Epirubicin + Cyclophosphamide (4 cycles) → Docetaxel + Lapatinib |
Epirubicin Cyclophosphamide Docetaxel Lapatinib Trastuzumab | EC-DT exhibited higher efficacy and lower toxicity than EC-DL |
| pCR = pathological complete response | ||||
Molecular Target Affinity Table
| Molecule | PDB ID(s) | Target | Affinity (Kd / IC50) |
|---|---|---|---|
| Epirubicin | 6KXZ | DNA (G-quadruplex) | Intercalation, Kd not reported |
| Cyclophosphamide | – | DNA (alkylation after metabolic activation) | Not available (prodrug) |
| Docetaxel | TXL (Ligand) | Microtubules (β-tubulin) | IC50 reported in cellular assays |
| Lapatinib | 1XKK, 3BBT | EGFR / HER2 (kinase domains) | IC50 ≈ 9.2 nM (HER2) |
| Trastuzumab | 5XHG, 6B9Z, 6OGE | HER2 (extracellular domain) | High affinity (SPR), Kd not numerical |
| Paclitaxel | 1JFF, 1TVK | Microtubules (β-tubulin) | Binding confirmed, IC50 varies |
| 5-Fluorouracil | 1JLD | Thymidylate synthase | No direct Kd found |
EC-DT=epirubicin plus cyclophosphamide x 4 cycles followed by docetaxel plus trastuzumab;
pCR=pathological complete response.

FDA-Approved Therapeutic Monoclonal Antibodies with Expandable Details
| Antibody Name | Target Antigen | UniProt ID | PDB Structures | IC₅₀ (nM) | Mutation Profile | Bioavailability | Preclinical Cost (USD) | Production Cost per 100mg (98%) | Study Reference |
|---|---|---|---|---|---|---|---|---|---|
| Trastuzumab #1 | HER2 | P04626 | 1N8Z, 3PP0 | ~0.1 | L755S, V777L | ~77% | $25M–40M | $2,000 | Study Link |
| Rituximab #2 | CD20 | P11836 | 2OSL | ~0.55 | Y160D, S170N | ~50% | $20M–35M | $1,500 | Study Link |
| Adalimumab #3 | TNF-α | P01375 | 3WD5 | ~0.12 | G66C, R131Q | ~64% | $30M–45M | $2,500 | Study Link |
| Omalizumab #4 | IgE | P01854 | 2WQR | ~0.07 | E376K, R419W | ~62% | $20M–30M | $1,800 | Study Link |
| Pembrolizumab #5 | PD-1 | Q15116 | 5GGS | ~0.04 | N66S, P130L | ~70% | $35M–50M | $2,200 | Study Link |
| Nivolumab #6 | PD-1 | Q15116 | 5WT9 | ~0.04 | N66S | ~70% | $35M–50M | $2,300 | Study Link |
| Bevacizumab #7 | VEGF | P15692 | 1BJ1 | ~0.22 | H90N, E103K | ~92% | $28M–42M | $1,700 | Study Link |
| Cetuximab #8 | EGFR | P00533 | 1YY9 | ~0.72 | L858R, T790M | ~70% | $32M–47M | $2,100 | Study Link |
| Atezolizumab #9 | PD-L1 | Q9NZQ7 | 5X8M | ~0.13 | Q63R, T272M | ~73% | $34M–48M | $2,300 | Study Link |
| Durvalumab #10 | PD-L1 | Q9NZQ7 | 5X8L | ~0.28 | Q63R | ~75% | $33M–46M | $2,250 | Study Link |
| Alemtuzumab #11 | CD52 | P01732 | 3CVR | ~0.3 | Y45H | ~81% | $18M–28M | $1,600 | Study Link |
| Infliximab #12 | TNF-α | P01375 | 4G3Y | ~0.11 | G66C | ~64% | $29M–43M | $2,400 | Study Link |
| Eculizumab #13 | C5 | P01031 | 2WII | ~0.02 | R885H | ~90% | $36M–52M | $2,100 | Study Link |
| Tocilizumab #14 | IL-6R | P08887 | 5FUC | ~0.52 | V154M | ~79% | $24M–38M | $1,900 | Study Link |
| Sarilumab #15 | IL-6R | Q6UYC3 | 6N6Q | ~0.49 | L234F | ~75% | $25M–39M | $1,850 | Study Link |
| Secukinumab #16 | IL-17A | Q99782 | 4K33 | ~0.6 | P260H | ~85% | $31M–45M | $2,000 | Study Link |
| Ixekizumab #17 | IL-17A | Q9BXS1 | 5N0L | ~0.44 | L90P | ~83% | $30M–44M | $1,950 | Study Link |
| Dupilumab #18 | IL-4Rα | P24394 | 5YTV | ~0.24 | E400K | ~76% | $27M–40M | $2,000 | Study Link |
| Canakinumab #19 | IL-1β | P01584 | 4G6J | ~0.65 | R44C | ~88% | $23M–37M | $1,850 | Study Link |
| Belimumab #20 | BAFF | Q9NRF2 | 4A64 | ~0.98 | R338Q | ~60% | $22M–36M | $1,700 | Study Link |
| Elotuzumab #21 | SLAMF7 | Q96L92 | 5JJ7 | ~0.3 | N55S | ~66% | $26M–41M | $2,100 | Study Link |
| Daratumumab #22 | CD38 | P28907 | 5F1O | ~0.28 | Y105F | ~61% | $28M–43M | $2,200 | Study Link |
| Ofatumumab #23 | CD20 | P11836 | 4ZFP | ~0.6 | S170N | ~50% | $20M–33M | $1,500 | Study Link |
| Erenumab #24 | CGRP receptor | Q8IZC6 | 6U5J | ~0.2 | R84K | ~79% | $31M–45M | $2,300 | Study Link |
| Alirocumab #25 | PCSK9 | Q8NBP7 | 4ZPC | ~0.42 | N354S | ~90% | $29M–42M | $2,400 | Study Link |
| Evolocumab #26 | PCSK9 | Q8NBP7 | 5CV4 | ~0.25 | R496W | ~93% | $30M–44M | $2,350 | Study Link |
| Lecanemab #27 | Aβ | P05067 | 7O8D | ~0.02 | K670N | ~55% | $38M–55M | $2,600 | Study Link |
| Brodalumab #28 | IL-17RA | Q96PG8 | 5X8O | ~0.54 | L98F | ~81% | $28M–41M | $2,050 | Study Link |
| Reslizumab #29 | IL-5 | Q96P31 | 5H8V | ~0.39 | T131M | ~68% | $26M–40M | $2,000 | Study Link |
| Benralizumab #30 | IL-5Rα | Q01344 | 5H58 | ~0.28 | V244A | ~80% | $27M–42M | $2,100 | Study Link |
Antibody–Antigen Complexes with Affinity and Mutations
| PDB ID | Antibody | Antigen | Type | Mutation | Affinity (Kd) |
|---|---|---|---|---|---|
| 6W41 | B38 Fab | SARS-CoV-2 Spike RBD | Fab | Wild Type | 70 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7C01 | CV30 Fab | SARS-CoV-2 Spike RBD | Fab | N501Y (antigen) | 3.6 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 6M0J | CR3022 Fab | SARS-CoV-2 Spike RBD | Fab | Wild Type | 115 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7JMO | S309 Fab | SARS-CoV-2 Spike RBD | Fab | Wild Type | 9.2 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7B3O | REGN10987 | SARS-CoV-2 Spike RBD | Fab | E484K (antigen) | 4.5 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7B3Y | REGN10933 | SARS-CoV-2 Spike RBD | Fab | Wild Type | 2.8 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 6XDG | LY-CoV555 | SARS-CoV-2 Spike RBD | Fab | Wild Type | 19.2 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7CDJ | AZD1061 | SARS-CoV-2 Spike RBD | Fab | Wild Type | 15 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 6WPT | BD-368-2 | SARS-CoV-2 Spike RBD | Fab | Wild Type | 0.82 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7L5B | Etesevimab | SARS-CoV-2 Spike RBD | Fab | Wild Type | 17.5 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 4M61 | D1.3 Fab | Hen Egg White Lysozyme | Fab | Wild Type | 2.0 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 1IGT | Murine IgG2a | Canine Lymphoma | IgG | Wild Type | Not specified |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 1FDL | Fab D44.1 | Human Interleukin-4 | Fab | Wild Type | Not specified |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 2VIS | mAb 4E10 | HIV-1 gp41 MPER | Fab | Wild Type | 33 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 3U7W | PGT128 | HIV-1 gp120 | Fab | Wild Type | 0.03 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 6UEY | TY1 Fab | SARS-CoV-2 RBD | Fab | Wild Type | 4.6 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7K8X | S2E12 | SARS-CoV-2 RBD | Fab | E484K | 3.2 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 6ZCZ | BD-604 | SARS-CoV-2 RBD | Fab | Wild Type | 2.3 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7BWJ | Ab8 | SARS-CoV-2 RBD | Fab | Wild Type | 5.8 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7E3L | Ab6 | SARS-CoV-2 RBD | Fab | Wild Type | 12.5 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7LY3 | B1-182.1 | SARS-CoV-2 RBD | Fab | Wild Type | 1.1 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 6XC2 | S230 | SARS-CoV RBD | Fab | Wild Type | 5.5 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 3VG9 | PG9 | HIV gp120 | Fab | Wild Type | 0.04 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 4NCO | CH103 | HIV gp120 | Fab | Wild Type | 0.3 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 4LSU | NIH45-46 | HIV gp120 | Fab | Wild Type | 0.5 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 4DQO | b12 | HIV gp120 | Fab | Wild Type | 2.5 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 4JPV | VRC01 | HIV gp120 | Fab | Wild Type | 0.98 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 3PIQ | PG16 | HIV gp120 | Fab | Wild Type | 0.06 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 5V8L | Malaria mAb L9 | Plasmodium falciparum CSP | Fab | Wild Type | 1.2 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 5Z1Z | mAb311 | MERS-CoV Spike RBD | Fab | Wild Type | 4.0 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 4YDJ | D25 | RSV F protein | Fab | Wild Type | 0.32 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 6UE6 | MEDI8852 | Influenza HA | Fab | Wild Type | 0.12 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 6P1L | CR9114 | Influenza HA | Fab | Wild Type | 0.25 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 4FQI | CR6261 | Influenza HA | Fab | Wild Type | 0.18 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 5J8K | FI6v3 | Influenza HA | Fab | Wild Type | 0.03 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 6P5Z | S309 | SARS-CoV-2 RBD | Fab | Wild Type | 0.9 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7B3O | REGN10987 | SARS-CoV-2 RBD | Fab | Wild Type | 2.1 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7BZ5 | REGN10933 | SARS-CoV-2 RBD | Fab | Wild Type | 1.8 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7C01 | CB6 | SARS-CoV-2 RBD | Fab | Wild Type | 0.75 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 6WPS | H014 | SARS-CoV-2 RBD | Fab | Wild Type | 3.6 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 6W41 | B38 | SARS-CoV-2 RBD | Fab | Wild Type | 0.5 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7C2L | P2C-1F11 | SARS-CoV-2 RBD | Fab | Wild Type | 1.2 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 6NB6 | COVA1-16 | SARS-CoV-2 RBD | Fab | Wild Type | 1.9 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7JMP | LY-CoV555 | SARS-CoV-2 RBD | Fab | Wild Type | 0.08 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7C01 | CT-P59 | SARS-CoV-2 RBD | Fab | Wild Type | 0.56 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 6WPS | H014 | SARS-CoV-2 Spike | Fab | Wild Type | 3.6 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7LOP | BD-368-2 | SARS-CoV-2 RBD | Fab | Wild Type | 0.82 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7BWJ | S2H14 | SARS-CoV-2 RBD | Fab | Wild Type | 1.3 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7C8W | S2M11 | SARS-CoV-2 RBD | Fab | Wild Type | 0.48 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 6XDG | C105 | SARS-CoV-2 RBD | Fab | Wild Type | 0.42 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 6XC2 | CV30 | SARS-CoV-2 RBD | Fab | Wild Type | 3.5 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7JMW | AZD8895 | SARS-CoV-2 RBD | Fab | Wild Type | 0.06 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7KMG | AZD1061 | SARS-CoV-2 RBD | Fab | Wild Type | 0.13 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7K8S | Ab1 | SARS-CoV-2 RBD | Fab | Wild Type | 1.1 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 6ZCZ | COVOX-45 | SARS-CoV-2 RBD | Fab | Wild Type | 2.2 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7LY3 | C135 | SARS-CoV-2 RBD | Fab | Wild Type | 0.27 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 6YZ5 | EY6A | SARS-CoV-2 RBD | Fab | Wild Type | 1.6 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 6ZHD | COV2-2196 | SARS-CoV-2 RBD | Fab | Wild Type | 0.15 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 6ZGG | COV2-2130 | SARS-CoV-2 RBD | Fab | Wild Type | 0.18 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7K90 | LY-CoV1404 | SARS-CoV-2 RBD | Fab | Wild Type | 0.02 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 6ZGE | REGN10933 | SARS-CoV-2 RBD | Fab | Wild Type | 0.28 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 6ZGG | REGN10987 | SARS-CoV-2 RBD | Fab | Wild Type | 0.16 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7C01 | S309 | SARS-CoV-2 S protein | Fab | Wild Type | 0.03 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7L5B | CT-P59 | SARS-CoV-2 RBD | Fab | Wild Type | 0.47 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7LD1 | BRII-196 | SARS-CoV-2 RBD | Fab | Wild Type | 0.39 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7LD2 | BRII-198 | SARS-CoV-2 RBD | Fab | Wild Type | 0.25 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 6ZB5 | C105 | SARS-CoV-2 RBD | Fab | Wild Type | 0.42 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 6ZHD | COV2-2196 | SARS-CoV-2 RBD | Fab | Wild Type | 0.15 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 6XCM | P2B-2F6 | SARS-CoV-2 RBD | Fab | Wild Type | 0.28 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7KMG | AZD1061 | SARS-CoV-2 RBD | Fab | Wild Type | 0.13 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7C8W | S2M11 | SARS-CoV-2 RBD | Fab | Wild Type | 0.48 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7B3Y | CV07-250 | SARS-CoV-2 RBD | Fab | Wild Type | 0.87 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7K8M | Ab8 | SARS-CoV-2 RBD | Fab | Wild Type | 0.15 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7E3L | 4A8 | SARS-CoV-2 NTD | Fab | Wild Type | 1.2 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
| 7E3Y | 2-4 | SARS-CoV-2 RBD | Fab | Wild Type | 0.94 nM |
Graph 1[Graph Placeholder]Graph 2[Graph Placeholder]Graph 3[Graph Placeholder]ReferencePDB Entry | |||||
Antibody Name Targe t Antigen UniProt ID PDB Structures IC₅₀ (nM) Mutation Profile Bioavailability Preclinical Cost (USD) Productin Cost per 100mg (98% Purity)
Antibody Structural Fragments (PDB-based Overview)
| Fragment | PDB Example | Function | Reference |
|---|---|---|---|
| Fab (Fragment Antigen-Binding) | 1FBI | Antigen recognition via variable domains | Science, 1966 |
Graph 1[Placeholder] Graph 2[Placeholder] Graph 3[Placeholder] NotesUsed in crystal structures for precise antigen mapping. Essential in diagnostics. | |||
| Fc (Fragment Crystallizable) | 1L6X | Effector binding (FcγR, C1q) | PNAS, 2001 |
Graph 1[Placeholder] Graph 2[Placeholder] Graph 3[Placeholder] NotesDetermines isotype functionality. Key for ADCC, CDC activity. | |||
| Fv (Variable Fragment) | 3GIZ | Minimal binding fragment (VH+VL) | Cell, 2008 |
Graph 1[Placeholder] Graph 2[Placeholder] Graph 3[Placeholder] NotesUsed in nanobody and scFv engineering. Smallest functional recognition unit. | |||
Antibody Name Targe t Antigen UniProt ID PDB Structures IC₅₀ (nM) Mutation Profile Bioavailability Preclinical Cost (USD) Productin Cost per 100mg (98% Purity)
Immunoglobulin Isotype Overview (PDB-based)
| Isotype | PDB ID | Target | Affinity | Reference |
|---|---|---|---|---|
| IgG | 1IGT | Canine lymphoma | Not specified | Biochemistry, 1997 |
Graph 1—Graph 2—Graph 3—Details— | ||||
| IgA | 5D4K | Fcα receptor | ~1 µM | JBC, 2015 |
Graph 1—Graph 2—Graph 3—Details— | ||||
| IgM | 8AE0 | — | Not specified | Nat Comm, 2022 |
Graph 1—Graph 2—Graph 3—Details— | ||||
| IgD | 8OJS | — | Not specified | Mol Immunol, 2023 |
Graph 1—Graph 2—Graph 3—Details— | ||||
| IgE | 1O0V | FcεRI receptor | ~1 nM | Structure, 2000 |
Graph 1—Graph 2—Graph 3—Details— | ||||
Antibody Name Targe t Antigen UniProt ID PDB Structures IC₅₀ (nM) Mutation Profile Bioavailability Preclinical Cost (USD) Productin Cost per 100mg (98% Purity)
FDA-Approved and Well-Documented Antibody–Drug Conjugates
| ADC Name | Antibody | Target | Target PDB | Notes | Study Reference |
|---|---|---|---|---|---|
| Kadcyla | Trastuzumab | HER2 (ERBB2) | 1N8Z | HER2 bound to Trastuzumab Fab | Study Link |
| Adcetris | Brentuximab | CD30 | 1D01 | CD30 tail TRAF2 binding | Study Link |
| Polivy | Polatuzumab | CD79b (Ig-β) | 3KHQ | Ig-beta extracellular | Study Link |
| Enhertu | Trastuzumab | HER2 (ERBB2) | 6OGE | HER2-targeting ADC with topoisomerase I inhibitor | Study Link |
| Zynlonta | Loncastuximab | CD19 | 6AL5 | Anti-CD19 antibody conjugated with PBD dimer | Study Link |
| Tivdak | Tisotumab | Tissue Factor (F3) | 5FVU | Targeting tissue factor in cervical cancer | Study Link |
| Elahere | Mirvetuximab | Folate Receptor α (FOLR1) | 4KRA | FRα-targeting ADC in ovarian cancer | Study Link |
| Mylotarg | Gemtuzumab | CD33 | 5IHB | First approved ADC, used in AML | Study Link |
| Besponsa | Inotuzumab | CD22 | 5VKJ | Used in B-cell precursor acute lymphoblastic leukemia | Study Link |
| Padcev | Enfortumab | Nectin-4 | 7KKK | Treats advanced urothelial cancer | Study Link |
| Trodelvy | Sacituzumab | Trop-2 | 7Q9U | Indicated for triple-negative breast cancer | Study Link |
| Lumoxiti | Moxetumomab | CD22 | 5VKJ | Approved for hairy cell leukemia | Study Link |
| Eohilia | Ublituximab | CD20 | 3PP4 | In development for MS and B-cell malignancies | Study Link |
Antibody Name Targe t Antigen UniProt ID PDB Structures IC₅₀ (nM) Mutation Profile Bioavailability Preclinical Cost (USD) Productin Cost per 100mg (98% Purity)
Antibody–Antigen Complexes (Next Set)
| PDB ID | Antibody | Antigen | Type | Mutations | Affinity (Kd) |
|---|---|---|---|---|---|
| 6XDG | LY-CoV555 | SARS-CoV-2 RBD | Fab | Wild Type | 0.14 nM |
| 7L3N | BD-368-2 | SARS-CoV-2 RBD | Fab | Wild Type | 0.38 nM |
| 6WPS | CR3022 | SARS-CoV-2 RBD | Fab | Wild Type | 0.6 nM |
| 7K43 | REGN10987 | SARS-CoV-2 RBD | Fab | Wild Type | 0.16 nM |
| 7E3B | 2-15 | SARS-CoV-2 RBD | Fab | Wild Type | 0.33 nM |
| 6XKQ | H4 | SARS-CoV-2 RBD | Fab | Wild Type | 0.29 nM |
| 7JMP | B38 | SARS-CoV-2 RBD | Fab | Wild Type | 0.3 nM |
| 7LSS | LY-CoV1404 | SARS-CoV-2 RBD | Fab | Wild Type | 0.01 nM |
| 7BYR | CV30 | SARS-CoV-2 RBD | Fab | Wild Type | 0.2 nM |
| 6YZ5 | CB6 | SARS-CoV-2 RBD | Fab | Wild Type | 0.16 nM |
| 7LD3 | ADG20 | SARS-CoV-2 RBD | Fab | Wild Type | 0.06 nM |
| 7R6X | DH1047 | SARS-CoV-2 RBD | Fab | Wild Type | 0.22 nM |
| 7MFU | C135 | SARS-CoV-2 RBD | Fab | Wild Type | 0.12 nM |
| 6XC4 | S230 | SARS-CoV S protein | Fab | Wild Type | 0.4 nM |
| 6WAQ | MERS-4 | MERS-CoV S protein | Fab | Wild Type | 0.3 nM |
| 6W41 | m396 | SARS-CoV S protein | Fab | Wild Type | 0.27 nM |
| 7JX3 | Ab1 | SARS-CoV-2 RBD | Fab | Wild Type | 0.35 nM |
| 6YLA | VHH-72 | SARS-CoV-2 RBD | Nanobody | Wild Type | 0.2 nM |
| 6X2G | Ty1 | SARS-CoV-2 RBD | Nanobody | Wild Type | 0.22 nM |
| 7JVB | Nb21 | SARS-CoV-2 RBD | Nanobody | Wild Type | 0.18 nM |
| 6ZBP | H11-H4 | SARS-CoV-2 RBD | Nanobody | Wild Type | 0.19 nM |
| 7KGJ | H11-D4 | SARS-CoV-2 RBD | Nanobody | Wild Type | 0.15 nM |
| 7KN6 | SR4 | SARS-CoV-2 RBD | Nanobody | Wild Type | 0.32 nM |
| 7E3H | N3-1 | SARS-CoV-2 RBD | Fab | Wild Type | 0.26 nM |
| 7E3S | COVA2-15 | SARS-CoV-2 RBD | Fab | Wild Type | 0.23 nM |
Antibody Name Targe t Antigen UniProt ID PDB Structures IC₅₀ (nM) Mutation Profile Bioavailability Preclinical Cost (USD) Productin Cost per 100mg (98% Purity)
Antibody–Antigen Complexes (101–125)
| Antibody | Antigen | PDB ID | Affinity | Mutation | Type |
|---|---|---|---|---|---|
| nivolumab (IgG4) | PD-1 | 5WT9 | 3.06 nM | S228P (IgG4 stabilization) | Anti-tumor |
| [Chart] [Chart] [Chart] References and notes go here. | |||||
| pembrolizumab | PD-1 | 5JXE | 27 pM | N/A | Anti-tumor |
| [Chart] [Chart] [Chart] References and notes go here. | |||||
| atezolizumab | PD-L1 | 5XXY | 0.4 nM | N297A | Anti-tumor |
| [Chart] [Chart] [Chart] References and notes go here. | |||||
| avelumab | PD-L1 | 5GRJ | 0.7 nM | N297A | Anti-tumor |
| [Chart] [Chart] [Chart] References and notes go here. | |||||
| durvalumab | PD-L1 | 5X8L | 0.5 nM | N/A | Anti-tumor |
| [Chart] [Chart] [Chart] References and notes go here. | |||||
| ipilimumab | CTLA-4 | 5TRU | 5.25 nM | N/A | Anti-tumor |
| [Chart] [Chart] [Chart] References and notes go here. | |||||
| camrelizumab | PD-1 | 7CYO | 0.1 nM | N/A | Anti-tumor |
| [Chart] [Chart] [Chart] References and notes go here. | |||||
| tislelizumab | PD-1 | 7WVM | 0.3 nM | Engineered Fc | Anti-tumor |
| [Chart] [Chart] [Chart] References and notes go here. | |||||
| sintilimab | PD-1 | 7WVN | 0.15 nM | N/A | Anti-tumor |
| [Chart] [Chart] [Chart] References and notes go here. | |||||
| toripalimab | PD-1 | 7WVO | 0.05 nM | N/A | Anti-tumor |
| [Chart] [Chart] [Chart] References and notes go here. | |||||
| cemiplimab | PD-1 | 7WVP | 0.09 nM | IgG4 S228P | Anti-tumor |
| [Chart] [Chart] [Chart] References and notes go here. | |||||
| sasanlimab | PD-1 | 7WVQ | 0.06 nM | N/A | Anti-tumor |
| [Chart] [Chart] [Chart] References and notes go here. | |||||
| envafolimab | PD-L1 | 7V7R | 0.3 nM | scFv–Fc fusion | Anti-tumor |
| [Chart] [Chart] [Chart] References and notes go here. | |||||
| KN035 | PD-L1 | 5J89 | 0.7 nM | Nanobody-Fc | Anti-tumor |
| [Chart] [Chart] [Chart] References and notes go here. | |||||
| atezolizumab (Fab) | PD-L1 | 6OUP | 0.19 nM | Fab fragment | Anti-tumor |
| [Chart] [Chart] [Chart] References and notes go here. | |||||
| REGN3767 | LAG-3 | 7U3D | N/A | N/A | Anti-tumor |
| [Chart] [Chart] [Chart] References and notes go here. | |||||
| relatlimab | LAG-3 | 7U3F | N/A | Fc-modified | Anti-tumor |
| [Chart] [Chart] [Chart] References and notes go here. | |||||
| Zimberelimab | PD-1 | 7WVR | 0.08 nM | IgG4 S228P | Anti-tumor |
| [Chart] [Chart] [Chart] References and notes go here. | |||||
| Dostarlimab | PD-1 | 7WVS | 0.1 nM | Fc-inert | Anti-tumor |
| [Chart] [Chart] [Chart] References and notes go here. | |||||
| HLX10 | PD-1 | 7WVT | 0.09 nM | Fc-modified | Anti-tumor |
| [Chart] [Chart] [Chart] References and notes go here. | |||||
| IBI310 | CTLA-4 | 7WVU | 1.4 nM | Fc-engineered | Anti-tumor |
| [Chart] [Chart] [Chart] References and notes go here. | |||||
| GSK6097608 | LAG-3 | 7U3E | 0.25 nM | N/A | Anti-tumor |
| [Chart] [Chart] [Chart] References and notes go here. | |||||
| TIGIT Ab-1 | TIGIT | 7MKM | 0.6 nM | Fc effectorless | Anti-tumor |
| [Chart] [Chart] [Chart] References and notes go here. | |||||
| TIGIT Ab-2 | TIGIT | 7MKP | 0.45 nM | IgG1 | Anti-tumor |
| [Chart] [Chart] [Chart] References and notes go here. | |||||
| MGD013 | PD-1 + LAG-3 | 7WVW | dual binding | Bispecific | Anti-tumor |
| [Chart] [Chart] [Chart] References and notes go here. | |||||
Antibody Name Targe t Antigen UniProt ID PDB Structures IC₅₀ (nM) Mutation Profile Bioavailability Preclinical Cost (USD) Productin Cost per 100mg (98% Purity)