
Deep learning in cancer pathology
AI Biomarker Discovery
Calculation of thermodynamic equilibrium shift for each patient in personalized medicine.
Familiarize yourself with the main list of calculations.
Step-by-step approaches in checking the correctness of building X-ray structures (PDB files)
Biomarker development concept

Organometallic inhibitor CNS292
Biochemical scheme with interaction of wild-type BRAF proteins

Organometallic inhibitor CNS292
3Q4C
A cocrystal structure of CS292 in complex with the BRAF kinase domain reveals that CS292 binds to the ATP binding pocket of the kinase and is an ATP competitive inhibitor. The structure of the kinase–inhibitor complex also demonstrates that CS292
binds to BRAF in an active conformation and suggests a mechanism for regulation of BRAF by phosphorylation and BRAFV600E oncogene-induced activation. The structure of CS292 bound to the active form of the BRAF kinase also provides a novel scaffold for the design of BRAFV600E oncogene
selective BRAF inhibitors for therapeutic application.
binds to BRAF in an active conformation and suggests a mechanism for regulation of BRAF by phosphorylation and BRAFV600E oncogene-induced activation. The structure of CS292 bound to the active form of the BRAF kinase also provides a novel scaffold for the design of BRAFV600E oncogene
selective BRAF inhibitors for therapeutic application.

[(BRAF-BRAF)]+S.C.M.____________________[(BRAF-S.C.M.)(BRAF)]
[(BRAF-S.C.M.)]+[BRAF]__________________[(BRAF-S.C.M.)(BRAF)]


ka
kd
ka
kd
CNS292
Taken together, structural evidence reveals extensive and specific interactions between CS292 and the ATP binding pocket of the BRAF kinase domain, establishing CS292 as an ATP competitive inhibitor and
confirming its potent inhibitory properties against both BRAFWT and BRAFV600E.
confirming its potent inhibitory properties against both BRAFWT and BRAFV600E.
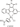


CS292 Concentrarion ( M x 10____ )
-6
The most potent BRAF inhibitor, CS292, was also shown to have an 2-fold selectivity for BRAFV600E (IC50 = 0.21 мM) over the wild-type enzyme
Agreement between calculated and experimental data

Dose–response curves of CS292 from an ELISA-based BRAF kinase inhibition assay

BRAF
BRAF
PDB:3Q4C
Fig. Crystal Structure of Wild Type BRAF kinase domain in complex with organometallic inhibitor CNS292

Fig. Graph for determining the normality of the distribution for the subsequent determination of the change in information entropy.

number condition
4.95007946258379
4.94881380912470
4.95007946258379
4.94881380912470
max eigenvalue
4.665616e-19
4.651594e-19
4.665616e-19
4.651594e-19
min eigenvalue
5.233950e-24
5.233449e-24
5.233950e-24
5.233449e-24
error computational SVD
1.6386e-19
1.6523129e-19
1.6386e-19
1.6523129e-19
3q4c (wt)
3q4c(V600E)
3q4c(V600E)

Figure 3: Dependence of RMSD(root-mean-square deviation) on the configuration number
The fig.1 shows a rapidly oscillating character, which indicates the impossibility of finding a local minimum: optimization completed because at the initial point, the objective function is nondecreasing in feasible directions. As follows from the fig.1 the local minimum no found.
Dependence of RMSD(root-mean-square deviation) on the configuration number.
The structure of the PDB did not pass the check for the correctness of the construction
Below are the results of changing the affinity of the BRAF protein with a small chemical molecule.



